Our bank conducts DNA barcoding tests to identify the animal species of origin of cells. This test amplifies the DNA barcode (a short nucleotide sequence of a specific gene region)*1 by PCR and then performs DNA sequencing of the PCR products.
By comparing the sequencing results with the nucleotide sequences of various animals registered in public databases*2, we can identify the animal species. With the advancement of DNA sequencing technology, DNA barcode sequencing has been performed on various animal species, and the results are now registered in public databases, making it possible to apply this method to species identification.
*1 DNA barcode used for identifying animal species of origin of the cell lines in our cell bank.
Our bank uses the mitochondrial Cytochrome c Oxidase subunit I (COI) gene.
If identification of the animal species cannot be achieved with the COI gene alone, we also analyze other gene sequences from animal species for which data are reported, such as Cytochrome b (Cytb) and 16S rRNA.
*2 Public Databases for identifying animal species of origin
Barcode of Life Data Systems(BOLD)
http://www.boldsystems.org/
National Center for Biotechnology Information – BLAST (Basic Local Alignment Search Tool)
https://blast.ncbi.nlm.nih.gov/Blast.cgi
1. Test Procedure
① DNA Extraction → ② PCR amplification of DNA barcode region → ③ Confirmation by gel electrophoresis → ④ Purification of amplified product → ⑤ DNA Sequencing → ⑥ Analysis using public databases
2. Items to Prepare
(1) Sample: Genomic DNA extracted from cells
(2) Equipment: Thermal cycler, such as Applied Biosystems® Veriti® Thermal Cycler
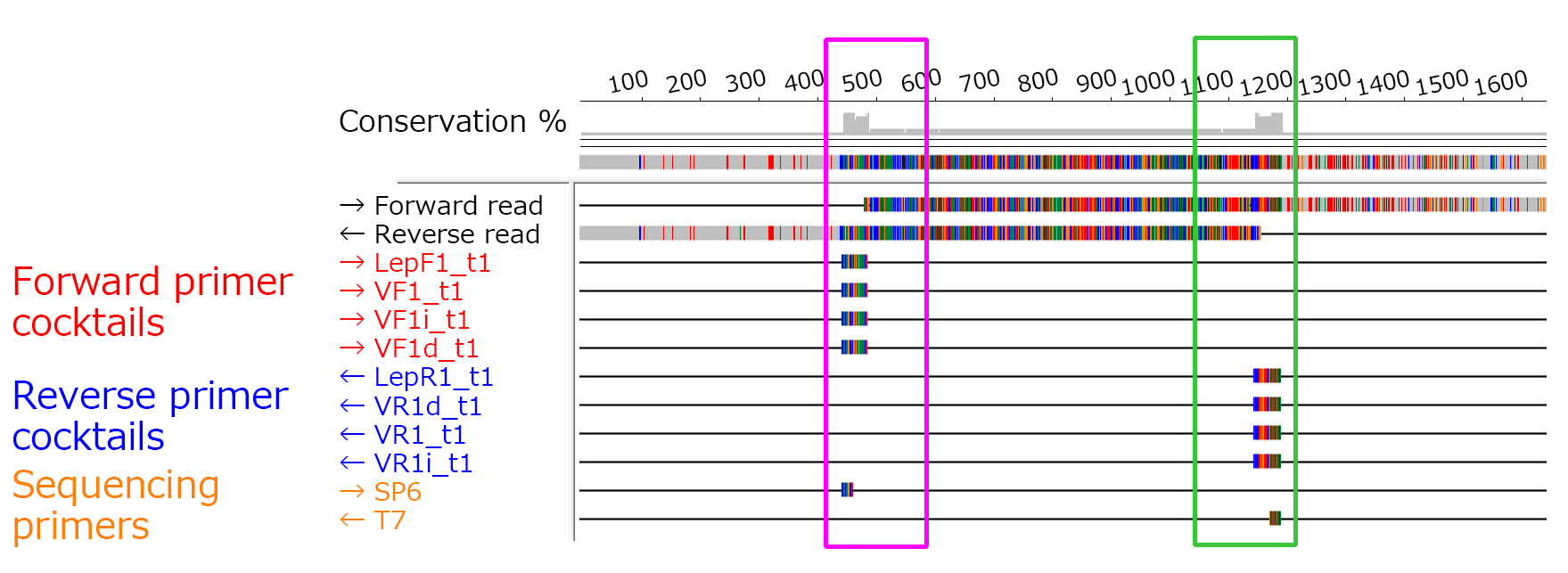
(3) PCR Primers
A) Cocktail primers for amplifying the COI gene
Universal primer cocktails *These primers amplify a wide range of animal species.
Mammalian primer mix for routine COI barcoding (Ivanova et al, 2007. Mol Ecol Notes 7:544-548)
*In the original method, M13 sequences were added for sequencing; however, in the method of our bank SP6 and T7 sequences were added for sequencing to the forward and reverse primers, respectively.
C_VF1LFt1_SP6 Forward primer cocktails (mixed in a 1:1:1:3 ratio)
C_VR1LRt1_T7 Reverse primer cocktails (mixed in a 1:1:1:3 ratio)
B) Species-specific primers for amplifying the COI gene *These primers are designed to amplify specific animal species.
In some cases, the cocktail primers in A) may not work well for amplification or sequencing. If the specific COI gene primers set or the COI gene sequence information of the target animal species is publicly available, our bank will construct species-specific primers and use them for amplification.
C) Primers for amplifying DNA barcodes other than the COI gene
The availability of COI gene database registration varies among animal species. Especially for “Asian endemic species” or subspecies of the same animal species from different habitats, there may be none or very few database registration, making identification difficult. In such cases, we search for alternative methods using other species- or subspecies-specific genes in medical reference databases, and construct primers for amplification.
D) Sequencing Primers
If the gene amplification primers in A) to C) are added with universal sequences such as SP6 or T7, these universal sequences are used as sequencing primers. If no universal sequences are added, the same primers used for gene amplification are used for sequencing.
3. Method
*The following describes the procedure using “A) Cocktail primers for amplifying the COI gene.” For PCR conditions using other primers, please refer to the references.
(1) PCR reaction mixture (for 1 reaction)
*If band excision and purification are required before sequencing, prepare multiple reactions.
| 10X Ex Taq Buffer | 5.0 μL |
| dNTP Mixture (2.5 mM each) | 4.0 μL |
| Forward Primer [C_VF1LFt1_SP6] (10 μM) | 0.5 μL |
| Reverse Primer [C_VR1LRt1_T7] (10 μM) | 0.5 μL |
| TaKaRa Ex Taq HS (5 units/μL) | 0.25 μL |
| DDW | 37.75 μL |
|
|
|
| Total | 48.0 μL |
(2) Dispense 48 μL of PCR reaction mixture into a PCR tube, and add 20ng (10ng/μL, 2μL) of template DNA from the sample to each tube.
(3) PCR Cycle

(4) Confirmation by Gel Electrophoresis Example results (using 2% agarose gel)
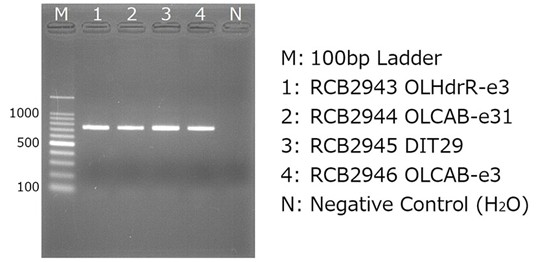
(5) Purification and absorbance measurement of amplified products
Purify the amplified product using commercially available PCR purification kits or gel extraction kits. Next, measure the absorbance of the amplified product and adjust it to the required concentration for sequencing analysis.
QIAquick PCR Purification Kit (50 / 250)(Cat No. 28104 / 28106)
QIAquick Gel Extraction Kit (50 / 250)(Cat No. 28704 / 28706) etc.
(6) Perform DNA sequencing analysis using a capillary sequencer with the Sanger method.
Once the sequence data and chromatogram are obtained, clean the poorly read regions at the “start” and “end” to create a consensus sequence.
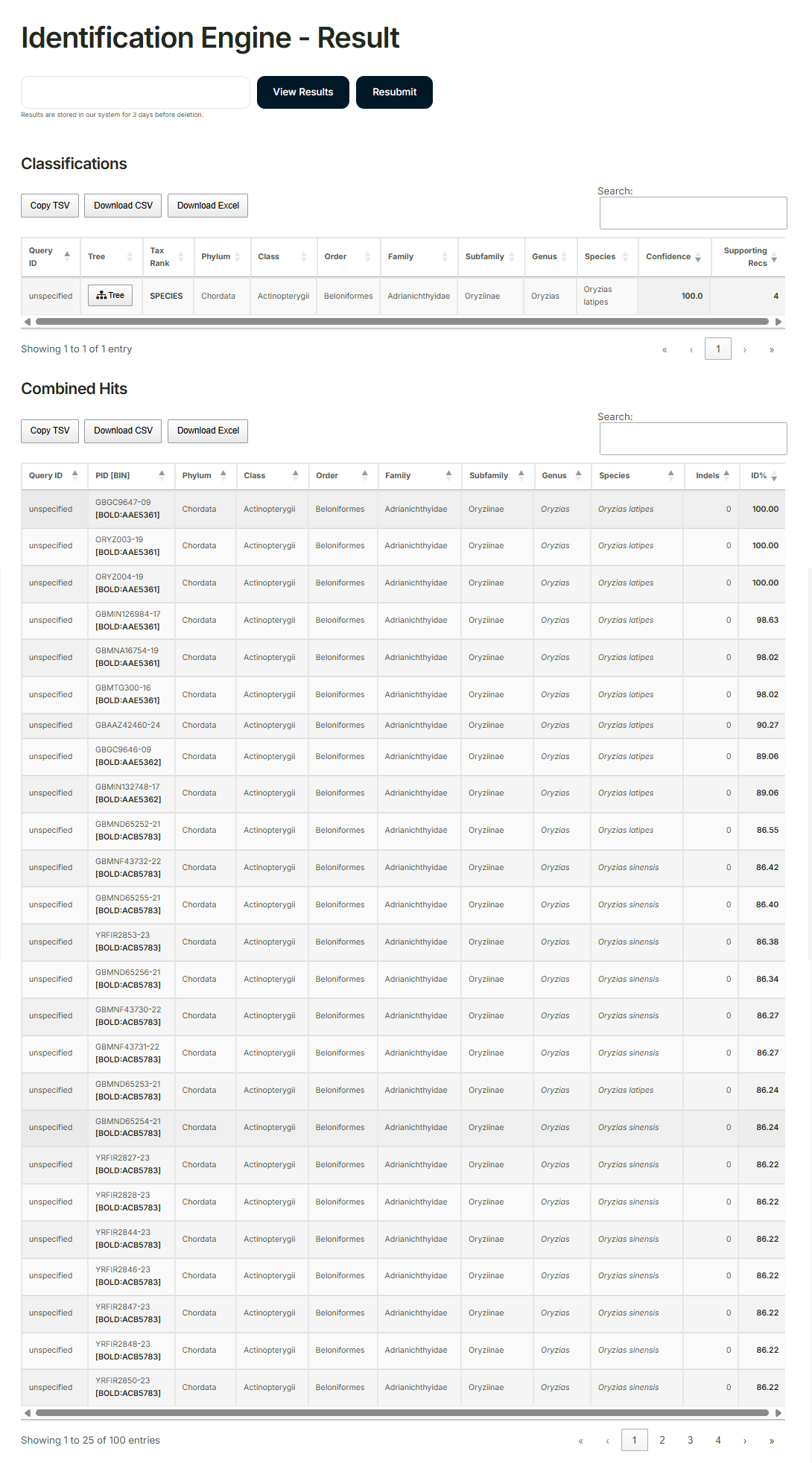
(7) Result analysis
Enter the consensus sequence into the “Barcode of Life Data Systems (BOLD)” for analysis.
Example of analysis results: RCB2943 OLHdrR-e3
https://cellbank.brc.riken.jp/cell_bank/CellInfo/?cellNo=RCB2943&lang=en
Identified species: Oryzias latipes 100%

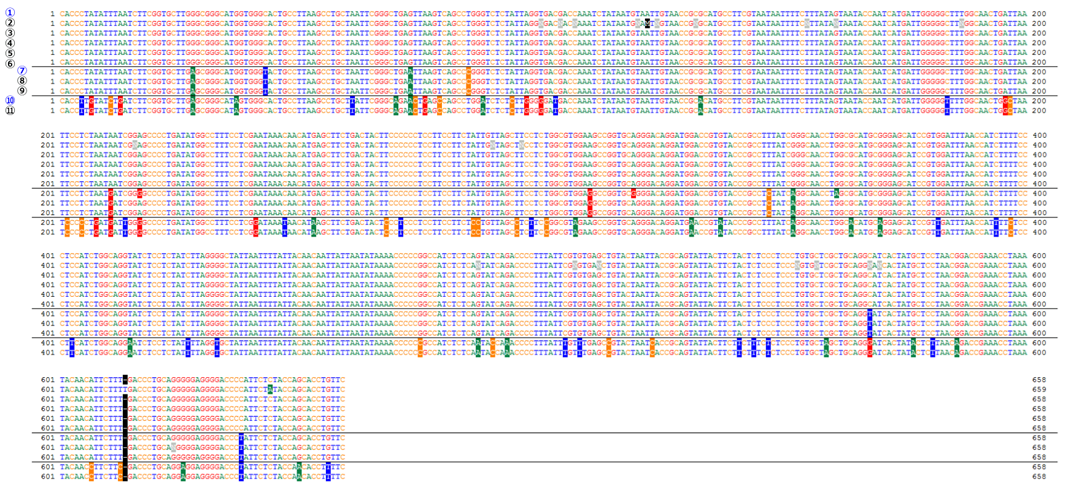
Comparison of COI gene sequences (cell lines derived from Oryzias latipes)
| ① Oryzias latipes COI gene, strain: Hd-rR https://www.ncbi.nlm.nih.gov/nuccore/AB498065 |
⑦ Oryzias latipes COI gene https://www.ncbi.nlm.nih.gov/nuccore/NC_004387 |
| ② RCB0187 OLHE-131 Lot.1 :OLHE-131 | ⑧ RCB0184 OLF-136 Lot.1 :OLF-136 |
| ③ RCB2943 OLHdrR-e3 Lot.1 :OLHdrR-e3 | ⑨ RCB0188 OLME-104 Lot.2 :OLME-104 |
| ④ RCB2944 OLCAB-e31 Lot.1 :OLCAB-e31 | ⑩ Oryzias latipesCOI gene, strain: HNI https://www.ncbi.nlm.nih.gov/nuccore/AB498066 |
| ⑤ RCB2945 DIT29 Lot.1 :DIT29 | ⑪ RCB2942 OLHNI-2 Lot.1 :OLHNI-2 |
| ⑥ RCB2946 OLCAB-e3 Lot.1 :OLCAB-e3 |
4. Interpretation of the result
If the analysis result from BOLD’s “Species Level Barcode Records” shows a match rate of 95% or higher and the species matches the deposited information, it will be considered a pass.
There are various cases depending on the animal species, such as “low match rate,” “no or few database registration,” “high match rate but unable to identify the subspecies,” “species name changed or reclassified due to new findings,” or “disagreement on the species name.” If identification cannot be achieved with the COI gene alone, alternative gene analysis methods will be considered, and a comprehensive judgment will be made.
5. References
Almeida, J.L., Cole, K.D., and Plant, A.L.
Standards for Cell Line Authentication and Beyond.
PLoS Biol 14: e1002476 (2016).
Hebert PD, Cywinska A, Ball SL, deWaard JR.
Biological identifications through DNA barcodes.
Proc Biol Sci. 2003 Feb 7;270(1512):313-21. doi:10.1098/rspb.2002.2218. PMID: 12614582; PMCID: PMC1691236.




