Mycoplasma Test
Mycoplasma is a microscopic organism and its presence often goes unnoticed even if it contaminates cultured cells or culture media. However, it is known that Mycoplasma contamination affects almost all metabolic reactions, cell characteristics, and proliferation within the cells. There is also the possibility that Mycoplasma contamination could spread unnoticed in the cell culture environment, making it desirable to perform Mycoplasma testing regularly.
Numerous methods have been developed for detecting Mycoplasma. In our bank, we conduct comprehensive testing by combining a DNA staining method (fluorescence dye detection) and a Nested-PCR method.
Before Mycoplasma Testing…
Samples containing antibiotics are less likely to detect Mycoplasma. To detect Mycoplasma contamination more accurately and with higher sensitivity, it is necessary to culture the cells in antibiotic-free media for more than 3 weeks.
Fluorescence Dye Detection Method (References 1–4)
Mycoplasma contamination in cultured cells is detected by staining Mycoplasma nuclear DNA with a fluorescent dye (Hoechst 33258) and observing the sample directly under a fluorescence microscope. Vero cells are used as indicator cells, i.e., co-cultured with the sample, to promote the growth of surviving Mycoplasma, allowing for high-sensitivity detection. Since Mycoplasma must be grown for detection, approximately 5 to 6 days are required for a more robust result. Furthermore, nuclear fragments derived from dead cells in both the sample and indicator cells may also be stained, requiring experience for accurate judgment.
Note: Human cord blood cells (HCB, CBF, C34) are not tested.
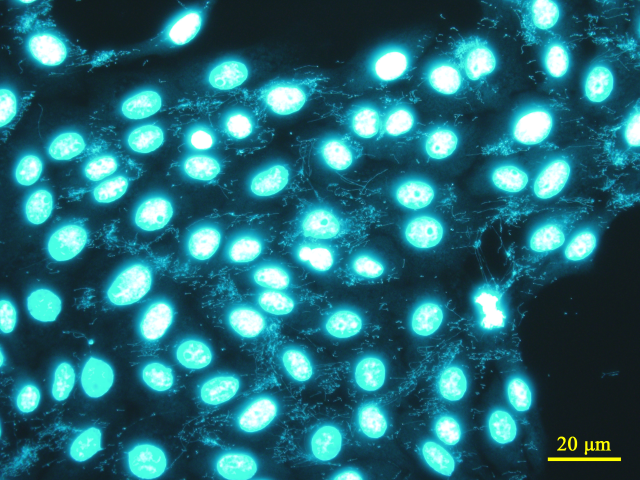
Fluorescence staining image of Vero cells contaminated with Mycoplasma (Fluorescent Dye: Hoechst 33258, Target: Nuclear DNA)
Nested-PCR Detection Method for Mycoplasma (References 5–8)
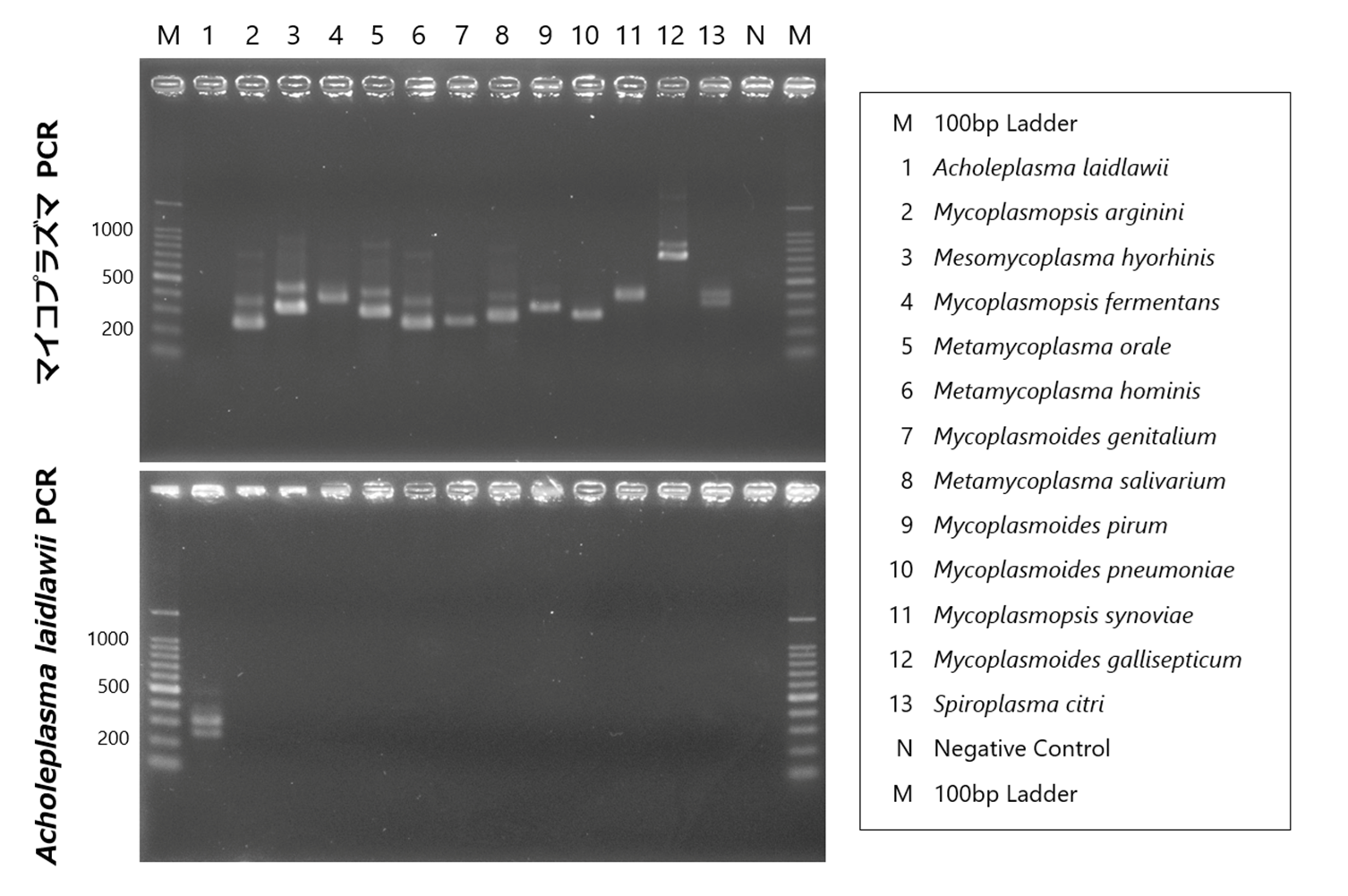
There are many types of Mycoplasma, and as shown in the figure below, species exhibit polymorphisms in the 16S-23S rRNA spacer region. By using primers that target the common sequences flanking this polymorphism, Mycoplasma can be detected using the Nested-PCR method. In addition to detecting the presence or absence of Mycoplasma, the amplified size can also give an indication of the species. This method allows for the rapid and simple detection of contamination by small amounts of Mycoplasma; however, there is a problem that not all types of Mycoplasma can be detected by this method alone.
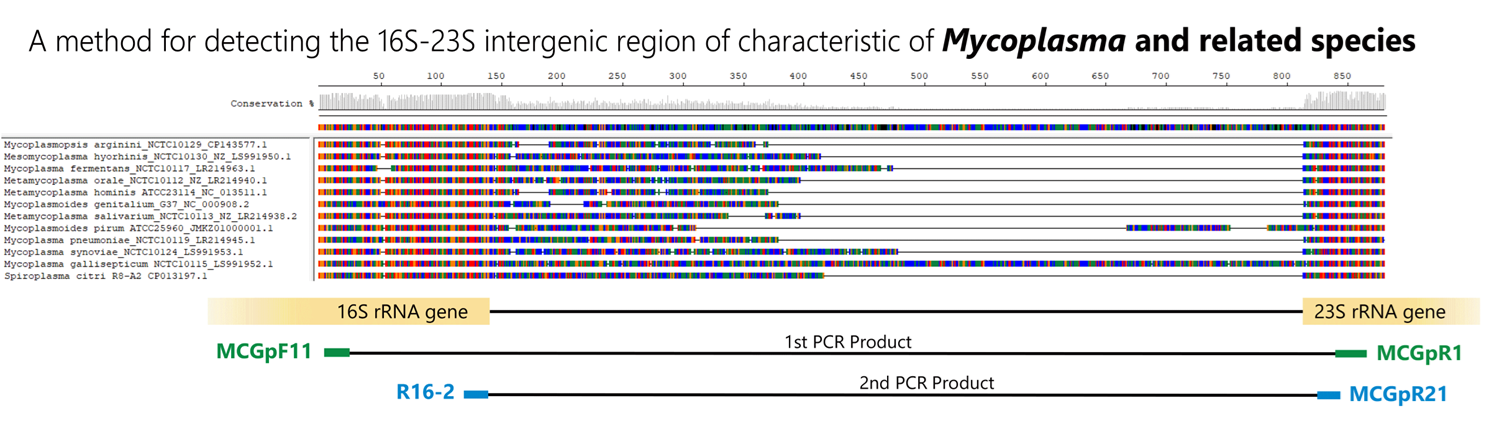
Control DNA detection and amplification sizes
Among the hundreds of Mycoplasma species and closely related species detectable by this PCR method, 12 species have been confirmed in our bank.
(* Mycoplasma species frequently infecting cultured cells)
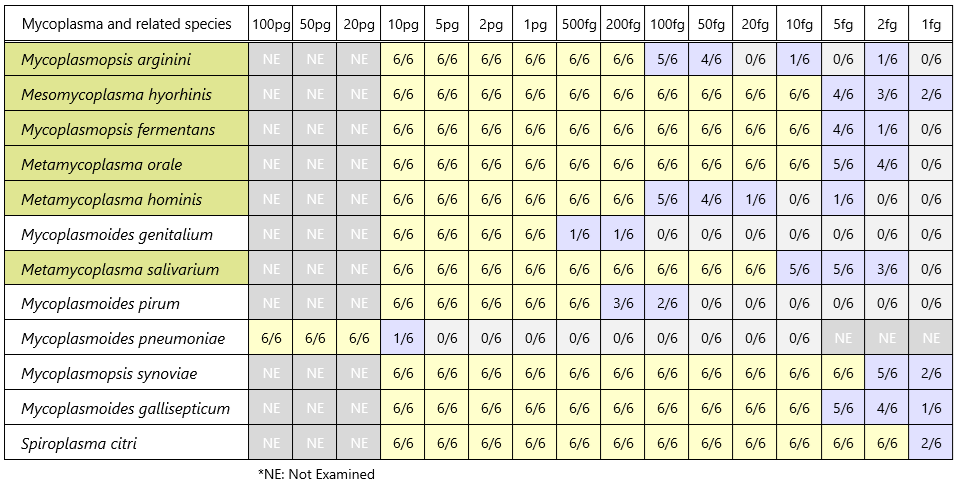
| Species | Main host | Strains | Source [Cat #] | 1st-step PCR product (bp) | 2nd-step PCR product (bp) |
| Mycoplasmopsis arginini | Bovine | NCTC10129 | Minerva Biolabs [51-0129] | 370 | 236 |
| Mesomycoplasma hyorhinis | Porcine | NCTC10130 | Minerva Biolabs [51-0130] | 448 | 314 |
| Mycoplasmopsis fermentans | Human | NCTC10117 | Minerva Biolabs [51-0117] | 492 | 366 |
| Metamycoplasma orale | Human | NCTC10112 | Minerva Biolabs [51-0112] | 424 | 290 |
| Metamycoplasma hominis | Human | ATCC 23114 | Minerva Biolabs [51-0111] | 370 | 236 |
| Mycoplasmoides genitalium | Human | ATCC 33530 | Minerva Biolabs [51-0195] | 390 | 252 |
| Metamycoplasma salivarium | Human | NCTC10113 | Minerva Biolabs [51-0113] | 403 | 269 |
| Mycoplasmoides pirum | Human | ATCC 25960 | ATCC [25960 D] | 459 | 323 |
| Mycoplasmoides pneumoniae | Human | NCTC10119 | Minerva Biolabs [51-0119] | 416 | 280 |
| Mycoplasmopsis synoviae | Avian | NCTC10124 | Minerva Biolabs [51-0124] | 496 | 362 |
| Mycoplasmoides gallisepticum | Avian | NCTC10115 | Minerva Biolabs [51-0115] | 846 | 710 |
| Spiroplasma citri | Insect, Plant | ATCC 27556 | Minerva Biolabs [51-0164] | 462 | 326 |
Detection sensitivity of control DNA (band detection count / PCR test count)
Nested-PCR detection method for A. laidlawii (Reference 9)
Using species-specific primers for Acholeplasma laidlawii, a species commonly infecting cultured cells, the 16S-23S rRNA spacer region is detected using the Nested-PCR method.
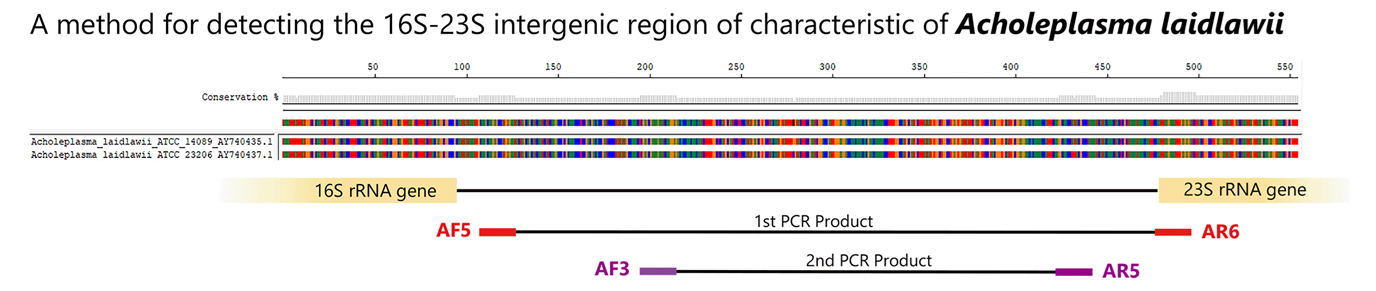
Control DNA detection and amplification sizes
| Species | Main host | Strains | Source [Cat #] | 1st-step PCR product (bp) | 2nd-step PCR product (bp) |
| Acholeplasma laidlawii | Bovine | NCTC10116 | Minerva Biolabs [51-0116] | 392 | 249 |
Detection sensitivity of control DNA (band detection count / PCR test count)

Positive control: DNA detection image
Depending on the amount of template added, both 1st-step and 2nd-step bands may appear.
References
- IFO Res. Comm., 13, 52-58 (1987)
- Bull. Jap. Fed. Culture Collections, 4, 9-15 (1988)
- 日本組織培養学会細胞バンク委員会報告書(平成2年3月)(Japanese only)
- 日本薬局方 バイオテクノロジー応用医薬品/生物期限由来医薬品の製造に用いる細胞基材に対するマイコプラズマ否定試験(第17改訂 参考情報 平成28年3月)(Japanese only)
- Res. Microbiol., 144, 489-493 (1993)
- Rapid Diagnosis of Mycoplasmas,227-232[eds.I.Kahane&A.Adoni](1993)
- 蛋白質核酸酵素40(15):2361-2368(1995) (Japanese only)
- Mol & Diag. Proced. In Mycoplasmology,vol.ⅡA7[eds.Razin&Tully](1996)
- 日本工業規格JIS K 3810-3:2003 (Japanese only)





